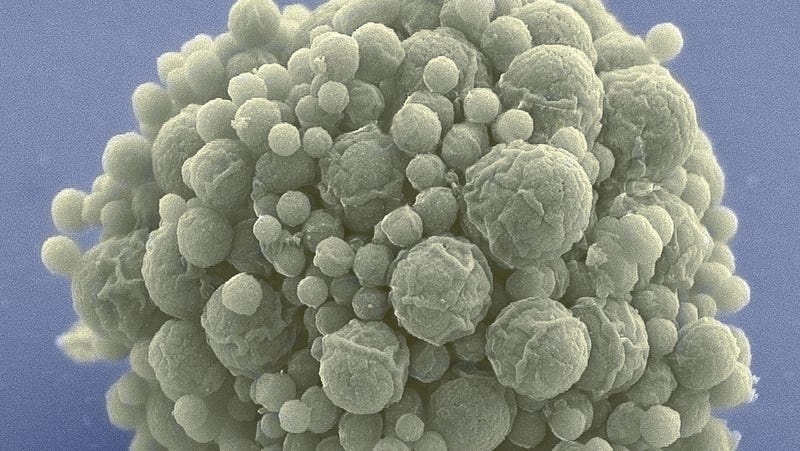
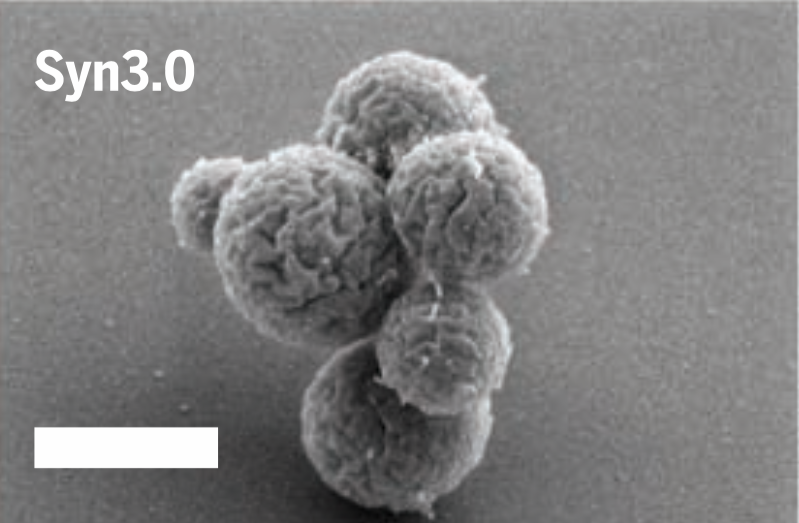
Behold syn3.0, a synthetic bacterial genome that’s smaller than anything found in nature. Biologists hope it will further our understanding of the fundamentals of life and inspire the creation of new synthetic life.
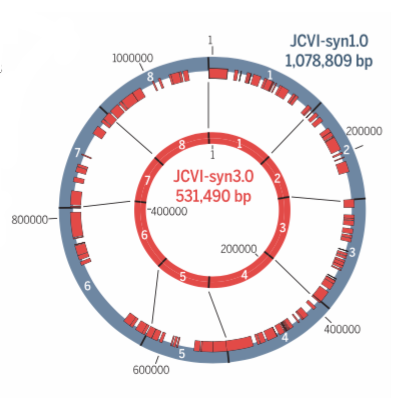
Dubbed JCVI-syn3.0 (or just syn3.0 for short), the new genome was designed and built by researchers from Synthetic Genomics and the J. Craig Venter Institute. It’s known as a “radically minimalist” genome because it has just 473 genes—the minimal number required for this bacterial cell to sustain the most basic functions of life, including reproduction. The details of this extraordinary achievement can now be found in the latest edition of Science.
It may sound like a strange endeavor, but there are some very good reasons for wanting to create a minimal genome. Evolutionary biologists will study it to better understand the processes under which complex organisms evolved from simpler ones. It could be used to explore the core functions of life, and to categorize essential genes within cells.
It also represents an important step in the nascent field of synthetic biology and the effort to build artificial organisms. Researchers could use this as a basic template to construct novel organisms with functions not seen in nature, including bacteria that can eat plastic and toxic waste, microorganisms that function like medicines inside the body, and biofuels comprised of organic components. More conceptually, this work could help astrobiologists predict what kinds of alien lifeforms might exist elsewhere in the cosmos.
This project, led by synthetic biologist Craig Venter and microbiologist Clyde Hutchison, dates back nearly 20 years to the very first sequencing of a bacterial genome. At the time, biologists began to speculate about a hypothetical minimal genome (HMG) for cells and other simple organisms. In 1995 it was theorized that a minimal bacterial cell genome would consist of 256 genes. Venter and his pals decided to test that assumption.
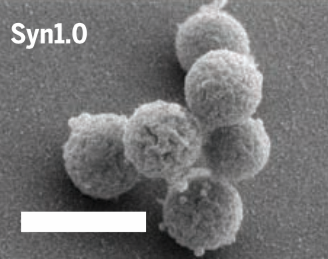
As Venter explained during a press briefing yesterday, “The only way to understand the minimal genome is to actually synthesize a genome.” Six years ago, his team accomplished that goal when it designed, constructed, and generated a self-replicating, synthetic bacterial cell derived from the Mycoplasma mycoides bacterium (so technically speaking the synthetic cell wasn’t designed from scratch). At the time, Venter described it as “the first species...to have its parents be a computer.” The experiment proved that genomes can be designed in a computer, then chemically created in the lab and transplanted into a recipient cell, and that these cells retain their capacity for self-replication.
The next step was to parse M. mycoides down to its essential elements. Venter thought it would take his team about a year, but the process turned out to be incredibly arduous, requiring four more years than initially planned. “Even with the sequence in hand, deciphering the operating system of the cell was a daunting task,” said Venter.
Early on in the process, every design failed, so they decided to try a new approach. The researchers constructed their synthetic cell from eight genetic segments, each of which could be tested independently within a genome that was seven-eighths complete.
They classified genes as being essential, quasi-essential (i.e. genes required for growth, but not absolutely required for life), or nonessential, and they established rules for removing genes from the genome design without disturbing expression of the remaining genes.
To deliberately knock out genetic function, the researchers inserted foreign genetic sequences, called transposons. This process was repeated until no more genes could be disrupted, and the genome was whittled down to the smallest size possible.
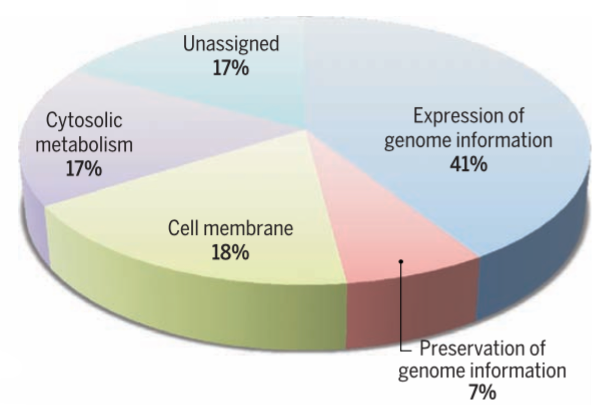
The researchers eventually came up with a new self-replicating cell. Of syn3.0's 473 genes, 438 encode proteins and 35 encode annotated RNAs. They were able to delete 428 genes from the starting total. The synthetic genome lacks all DNA-modifying and restrictions genes, and also genes responsible for encoding lipoproteins (proteins that transport fat or other lipids in the blood plasma). The researchers said it’s a “working approximation to a minimal cell,” with a genome “smaller than that of any autonomously replicating cell found in nature.” By comparison, the naturally occurring Mycoplasma genitalium bacterium has 525 genes.

During the process, however, the removal of some genes classified as “non-essential” caused unforeseen problems. Certain genes had to be retained in the minimal genome, despite having no known or obvious function. In total, the researchers were unable to determine the function of 149 genes, which is almost a third of the entire genome.
Based on analogous genes in other organisms, the researchers suspect that these genes have something to do with the encoding of universal proteins. But they’re not entirely sure.
This proved to be a rather eye-opening revelation, and the researchers said this is a high priority for the next stage of the project. It also shows how much there is still to know about whole-gene expression and design. As Venter put it, “The next stages are not going to be trivial.”
This lack of understanding shows how important it is to have a genome-centric view of the fundamentals of life, rather than a reductionist gene-specific view. “Life is more like an orchestra than a single piccolo player,” said Hutchison. “Sometimes you don’t discover essentiality until you remove a second [gene].”
In addition retaining some genes of unknown function, the researchers also had to include a significant number of quasi-essential genes required for robust growth, but not required for the basic functions of life. Technically speaking, the genome could have been stripped down even further, but cellular growth would have been so painstakingly slow as to render the cell useless for scientific experimentation. This shows that, during the miniaturization process, there’s a tradeoff between genome size and growth rate.
Syn3.0 represents a minimal genome for M. mycoides under ideal laboratory conditions. In all likelihood, it would not survive outside the lab. Any “adaptive” traits it might have had were removed. Also, other bacteria would undoubtedly have a different minimal genome, which is dependant on physical characteristics and environmental conditions. “Minimum is a relative term, based on your definition of traits and functions of a cell,” said Venter. “So if you want a cell to have [say] photosynthetic properties, there will be totally different set of genes to support those functions.”
At any rate, the point of the experiment was to create a baseline genome that scientists can use to study life, and to use as a “chassis” for adding new sets of genes. In theory, syn3.0 can be used to construct virtually any kind of cell with custom-built properties. As study co-author Dan Gibson explained, the long term vision is “to design and build synthetic organisms on demand,” and to “add functions and predict outcomes.” He said it could be used in many industrial applications, including medicine, biochemicals, biofuels, nutrition, and agriculture.
Venter and his colleagues are toying with the idea of launching a contest to see who can devise the most beneficial and innovative “gain of functions” for their minimal genome. In the meantime, they are filing patents, both on syn3.0 and the process used to create it.
[Science]